Example of asynchronous requests (v > 1.1)#
The scope of this example is to show how to request several products together so that internal resource usage is maximized
We extract the spectrum of the Crab in groups of ‘nscw’ science windows for each year from ‘start_year’ to ‘stop_year’ included
We use a token provided by the web interface to receive dedicated emails
We optionally show how to fit the spectra with a broken power law using xspec
[1]:
#A few input parameters
osa_version="OSA10.2"
source_name="Crab"
nscw=10
start_year=2004
end_year=2006
systematic_fraction = 0.01
token=''
Token authentication#
You can provide a valid token as explained in the ‘Authentication’ example or skip the following cell and continue anonymously
[2]:
import getpass
token = getpass.getpass('Insert the token')
Insert the token········
[3]:
# To know details of the token
import oda_api.token
oda_api.token.decode_oda_token(token)
[3]:
{'sub': 'Carlo.Ferrigno@unige.ch',
'email': 'Carlo.Ferrigno@unige.ch',
'name': 'cferrigno',
'roles': 'authenticated user, administrator, content manager, general, integral-private-qla, magic, unige-hpc-full, public-pool-hpc, antares, sdss',
'exp': 1631110932}
[3]:
#We hardcode a catalog for the Crab
api_cat={
"cat_frame": "fk5",
"cat_coord_units": "deg",
"cat_column_list": [
[0, 7],
["1A 0535+262", "Crab"],
[125.4826889038086, 1358.7255859375],
[84.72280883789062, 83.63166809082031],
[26.312734603881836, 22.016284942626953],
[-32768, -32768],
[2, 2],
[0, 0],
[0.0002800000074785203, 0.0002800000074785203]],
"cat_column_names": [
"meta_ID",
"src_names",
"significance",
"ra",
"dec",
"NEW_SOURCE",
"ISGRI_FLAG",
"FLAG",
"ERR_RAD"
],
"cat_column_descr":
[
["meta_ID", "<i8"],
["src_names", "<U11"],
["significance", "<f8"],
["ra", "<f8"],
["dec", "<f8"],
["NEW_SOURCE", "<i8"],
["ISGRI_FLAG", "<i8"],
["FLAG", "<i8"],
["ERR_RAD", "<f8"]
],
"cat_lat_name": "dec",
"cat_lon_name": "ra"
}
Let’s get some logging#
This is to help visualizing the progress.
WARNING is the default level
INFO writes some more information
DEBUG is maily for developers and issue tracking
[4]:
import logging
#default
#logging.getLogger().setLevel(logging.WARNING)
#slightly more verbose
logging.getLogger().setLevel(logging.INFO)
#all messages
#logging.getLogger().setLevel(logging.DEBUG)
logging.getLogger('oda_api').addHandler(logging.StreamHandler())
Different instances of the platform#
the general user will use the ‘production’ one, the other one is for internal testing
[5]:
import numpy as np
import json
import oda_api.api
import oda_api
from pkg_resources import parse_version
assert parse_version(oda_api.__version__) > parse_version("1.1.0")
def dispatcher(_oda_platform='production'):
disp = oda_api.api.DispatcherAPI(
url = {
'staging' : 'http://dispatcher.staging.internal.odahub.io',
'production': 'https://www.astro.unige.ch/mmoda/dispatch-data',
}[_oda_platform]
)
disp.get_instrument_description("isgri")
return disp
disp = dispatcher('production')
--------------
query_name: src_query
name: src_name, value: test, units: str,
name: RA, value: 0.0, units: deg,
name: DEC, value: 0.0, units: deg,
name: T1, value: 2001-12-11T00:00:00.000, units: None,
name: T2, value: 2001-12-11T00:00:00.000, units: None,
name: token, value: None, units: str,
--------------
query_name: isgri_parameters
name: user_catalog, value: None, units: str,
name: scw_list, value: [], units: names_list,
name: selected_catalog, value: None, units: str,
name: radius, value: 5.0, units: deg,
name: max_pointings, value: 50, units: None,
name: osa_version, value: None, units: str,
name: integral_data_rights, value: public, units: str,
name: E1_keV, value: 15.0, units: keV,
name: E2_keV, value: 40.0, units: keV,
--------------
query_name: isgri_image_query
product_name: isgri_image
name: detection_threshold, value: 0.0, units: sigma,
name: image_scale_min, value: None, units: None,
name: image_scale_max, value: None, units: None,
--------------
query_name: isgri_spectrum_query
product_name: isgri_spectrum
--------------
query_name: isgri_lc_query
product_name: isgri_lc
name: time_bin, value: 1000.0, units: sec,
--------------
query_name: spectral_fit_query
product_name: spectral_fit
name: xspec_model, value: powerlaw, units: str,
name: ph_file_name, value: , units: str,
name: arf_file_name, value: , units: str,
name: rmf_file_name, value: , units: str,
Here, we collect and spectra for each year in a random sample of nscw=10 science windows
We use the hard-coded catalog.
note that we make a loop and submit the jobs without waiting for their completion
at each loop, we test if they completed and we count how many have finished
we continue to poll the dispatcher for unfinished jobs and we terminate the loop when all are done
In this way, we let the platform optimize our requests
There will be a convenience function in future versions of oda_api for this purpose
[6]:
spectrum_results=[]
disp_by_ys = {}
data_by_ys = {}
par_dict = {"RA": "83.63166809082031",
"DEC": "22.016284942626953",
"radius": "10",
'instrument':'isgri',
'product': 'isgri_spectrum',
'osa_version' : osa_version,
'product_type': 'Real',
'max_pointings': nscw,
'selected_catalog' : json.dumps(api_cat)}
# Should you need to access private data, just add this option
#,"integral_data_rights": "all-private"}
if token != '':
par_dict.update({'token': token})
while True:
spectrum_results=[]
for year in range(start_year, end_year+1):
T1_utc='%4d-01-01T00:00:00.0'%year
T2_utc='%4d-12-31T23:59:59.0'%year
print(T1_utc,'-',T2_utc)
par_dict.update({'T1': T1_utc,
'T2': T2_utc})
if year >= 2016:
osa_version='OSA11.1'
else:
osa_version='OSA10.2'
#Just renaiming for a general dictionary key
ys = year
# We start one dipatcher for each job,
# they will run in parallel until products are ready
if ys not in disp_by_ys:
disp_by_ys[ys] = oda_api.api.DispatcherAPI(url=disp.url, wait=False) #Note the flag wait=False
_disp = disp_by_ys[ys]
data = data_by_ys.get(ys, None)
if data is None and not _disp.is_failed:
#We submit or we poll
if not _disp.is_submitted:
data = _disp.get_product(**par_dict)
else:
_disp.poll()
print("Is complete ", _disp.is_complete)
# We retrieve data
if not _disp.is_complete:
continue
else:
data = _disp.get_product(**par_dict)
data_by_ys[ys] = data
spectrum_results.append(data)
n_complete = len([ year for year, _disp in disp_by_ys.items() if _disp.is_complete ])
print(f"complete {n_complete} / {len(disp_by_ys)}")
if n_complete == len(disp_by_ys):
print("done!")
break
print("not done")
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
- waiting for remote response (since 2021-08-31 17:52:30), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
... query status prepared => submitted
... assigned job id: be2375ee6e04c3ef
| the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 22 seconds (23/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
non-waiting dispatcher: terminating
query not complete, please poll again later
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
- waiting for remote response (since 2021-08-31 17:52:53), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
... query status prepared => submitted
... assigned job id: 5ebc852ab66d5917
| the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 27 seconds (27/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
non-waiting dispatcher: terminating
query not complete, please poll again later
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
- waiting for remote response (since 2021-08-31 17:53:21), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
... query status prepared => submitted
... assigned job id: f9db3b072b6ffe3c
| the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 6 seconds (6.6/6.6); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
non-waiting dispatcher: terminating
query not complete, please poll again later
- waiting for remote response (since 2021-08-31 17:53:28), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
/ the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 63 seconds (14/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:53:33), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
/ the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 45 seconds (17/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:53:39), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
/ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 23 seconds (6.3/6.6); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:53:45), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
- the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 80 seconds (11/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:53:50), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
- the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 62 seconds (13/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:53:55), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
- the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 56 seconds (12/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:54:17), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
\ the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 130 seconds (14/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:54:40), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
\ the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 113 seconds (11/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:54:46), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
\ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 90 seconds (10/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:54:52), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
| the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 160 seconds (15/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:11), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
| the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 143 seconds (10/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:16), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
| the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 121 seconds (9.2/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:22), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
/ the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 177 seconds (13/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:28), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
/ the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 160 seconds (9.4/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:34), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
/ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 138 seconds (8.6/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:40), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
- the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 195 seconds (12/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:45), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
- the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 177 seconds (8.9/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:51), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
- the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 155 seconds (8.1/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:55:56), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
\ the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 227 seconds (13/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:56:18), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
\ the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 210 seconds (8.5/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:56:24), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
\ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 188 seconds (7.9/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:56:30), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
| the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 246 seconds (13/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:56:36), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
| the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 228 seconds (8.2/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:56:42), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
| the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 206 seconds (7.7/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:56:48), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
/ the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 263 seconds (12/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:56:53), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
/ the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 246 seconds (8/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:57:00), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
/ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 224 seconds (7.5/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:57:06), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
- the job is working remotely, please wait status=submitted job_id=be2375ee in 0 messages since 281 seconds (11/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:57:11), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
- the job is working remotely, please wait status=submitted job_id=5ebc852a in 0 messages since 262 seconds (7.7/27); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:57:16), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
- the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 240 seconds (7.3/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:57:22), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 0 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
... query status submitted => done
query COMPLETED SUCCESSFULLY (state done)
Is complete True
- waiting for remote response (since 2021-08-31 17:57:36), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
session: CHSJDFEI89QXQBX8 job: be2375ee6e04c3ef
... query status prepared => done
... assigned job id: be2375ee6e04c3ef
query COMPLETED SUCCESSFULLY (state done)
non-waiting dispatcher: terminating
- waiting for remote response (since 2021-08-31 17:57:51), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
... query status submitted => done
query COMPLETED SUCCESSFULLY (state done)
Is complete True
- waiting for remote response (since 2021-08-31 17:58:06), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
session: KITDUODRMMKQLUK9 job: 5ebc852ab66d5917
... query status prepared => done
... assigned job id: 5ebc852ab66d5917
query COMPLETED SUCCESSFULLY (state done)
non-waiting dispatcher: terminating
- waiting for remote response (since 2021-08-31 17:58:20), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
\ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 304 seconds (7.2/22); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:58:26), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 2 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
| the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 327 seconds (8.4/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:58:49), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 2 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
/ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 333 seconds (8.3/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:58:55), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 2 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
- the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 339 seconds (8.1/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:59:01), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 2 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
\ the job is working remotely, please wait status=submitted job_id=f9db3b07 in 0 messages since 345 seconds (7.9/23); in 0 SCW so far; nodes (0): 0 computed 0 restored
...
- waiting for remote response (since 2021-08-31 17:59:06), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
Is complete False
complete 2 / 3
not done
2004-01-01T00:00:00.0 - 2004-12-31T23:59:59.0
2005-01-01T00:00:00.0 - 2005-12-31T23:59:59.0
2006-01-01T00:00:00.0 - 2006-12-31T23:59:59.0
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
... query status submitted => done
query COMPLETED SUCCESSFULLY (state done)
Is complete True
- waiting for remote response (since 2021-08-31 17:59:21), please wait for https://www.astro.unige.ch/mmoda/dispatch-data/run_analysis
session: LZ9F8KABFTBZEW8Y job: f9db3b072b6ffe3c
... query status prepared => done
... assigned job id: f9db3b072b6ffe3c
query COMPLETED SUCCESSFULLY (state done)
non-waiting dispatcher: terminating
complete 3 / 3
done!
Elaboration example#
This part saves the spectra in fits files and updates some keywords
[7]:
from astropy.io import fits
# This part saves the spectra in fits files and updates some keywords
for year, data in data_by_ys.items():
print(year)
for ID,s in enumerate(data._p_list):
if (s.meta_data['src_name']==source_name):
if(s.meta_data['product']=='isgri_spectrum'):
ID_spec=ID
if(s.meta_data['product']=='isgri_arf'):
ID_arf=ID
if(s.meta_data['product']=='isgri_rmf'):
ID_rmf=ID
print(ID_spec, ID_arf, ID_rmf)
spec=data._p_list[ID_spec].data_unit[1].data
arf=data._p_list[ID_arf].data_unit[1].data
rmf=data._p_list[ID_rmf].data_unit[2].data
expos=data._p_list[0].data_unit[1].header['EXPOSURE']
name=source_name+'_'+str(year)
specname=name+'_spectrum.fits'
arfname=name+'_arf.fits.gz'
rmfname=name+'_rmf.fits.gz'
data._p_list[ID_spec].write_fits_file(specname)
data._p_list[ID_arf].write_fits_file(arfname)
data._p_list[ID_rmf].write_fits_file(rmfname)
hdul = fits.open(specname, mode='update')
hdul[1].header.set('EXPOSURE', expos)
hdul[1].header['RESPFILE']=rmfname
hdul[1].header['ANCRFILE']=arfname
hdul[1].data['SYS_ERR']=systematic_fraction
hdul.close()
2004
3 4 5
2005
3 4 5
2006
3 4 5
Elaboration 2#
If xspec is available, we make a fit of each spectrum
[8]:
try:
import xspec
import shutil
from IPython.display import Image
from IPython.display import display
xspec.Fit.statMethod = "chi"
#init dictionaries
fit_by_lt={}
model='cflux*bknpow'
xspec.AllModels.systematic=0.0
low_energies=[20]
freeze_pow_ebreak=1
for year in range(start_year,end_year+1):
for c_emin in low_energies: #np.linspace(17,40,5):
xspec.AllData.clear()
m1=xspec.Model(model)
specname=source_name+'_'+str(year)+'_spectrum.fits'
xspec.AllData(specname)
s = xspec.AllData(1)
isgri = xspec.AllModels(1)
print(m1.nParameters)
xspec.AllData.ignore('bad')
xspec.AllData.ignore('500.0-**')
ig="**-%.2f,500.-**"%c_emin
print("ISGRI ignore: "+ ig)
s.ignore(ig)
#Key for output
lt_key='%d_%.10lg'%(year, c_emin)
isgri.cflux.lg10Flux=-8
isgri.cflux.Emin=20.
isgri.cflux.Emax=80.
isgri.bknpower.norm = "1,-1"
isgri.bknpower.PhoIndx1 = "2.0,.01,1.,1.,3.,3."
isgri.bknpower.PhoIndx2 = "2.2,.01,1.,1.,3.,3."
isgri.bknpower.BreakE = "100,-1,20,20,300,300"
xspec.Fit.perform()
isgri.bknpower.BreakE.frozen = freeze_pow_ebreak > 0
xspec.Fit.perform()
max_chi=np.ceil(xspec.Fit.statistic / xspec.Fit.dof)
xspec.Fit.error("1.0 max %.1f 1-%d"%(max_chi,m1.nParameters))
fit_by_lt[lt_key]=dict(
emin=c_emin,
year=year,
chi2_red=xspec.Fit.statistic/xspec.Fit.dof,
chi2=xspec.Fit.statistic,
ndof=xspec.Fit.dof,
)
for i in range(1,m1.nParameters+1):
if (not isgri(i).frozen) and (not bool(isgri(i).link)):
#use the name plus position because there could be parameters with same name from multiple
#model components (e.g., several gaussians)
print(isgri(i).name, "%.2f"%(isgri(i).values[0]), isgri(i).frozen,bool(isgri(i).link) )
fit_by_lt[lt_key][isgri(i).name+"_%02d"%(i)]=[ isgri(i).values[0], isgri(i).error[0], isgri(i).error[1] ]
xspec.Plot.device="/png"
#xspec.Plot.addCommand("setplot en")
xspec.Plot.xAxis="keV"
xspec.Plot("ldata del")
xspec.Plot.device="/png"
fn="fit_%s.png"%lt_key
fit_by_lt[lt_key]['plot_fname'] = fn
shutil.move("pgplot.png_2", fn)
_=display(Image(filename=fn,format="png"))
except ImportError:
print("no problem!")
Default fit statistic is set to: Chi-Squared
This will apply to all current and newly loaded spectra.
7
Model systematic error set to 0
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/Off
Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 0.500000 frozen
2 1 cflux Emax keV 10.0000 frozen
3 1 cflux lg10Flux cgs -12.0000 +/- 0.0
4 2 bknpower PhoIndx1 1.00000 +/- 0.0
5 2 bknpower BreakE keV 5.00000 +/- 0.0
6 2 bknpower PhoIndx2 2.00000 +/- 0.0
7 2 bknpower norm 1.00000 +/- 0.0
________________________________________________________________________
Warning: RMF CHANTYPE keyword (PHA) is not consistent with that from spectrum (PI)
1 spectrum in use
Spectral Data File: Crab_2004_spectrum.fits Spectrum 1
Net count rate (cts/s) for Spectrum:1 -nanISGRI ignore: **-20.00,500.-**
+/- -nan
Assigned to Data Group 1 and Plot Group 1
Noticed Channels: 1-62
Telescope: INTEGRAL Instrument: IBIS Channel Type: PI
Exposure Time: 1.277e+04 sec
Using fit statistic: chi
Using Response (RMF) File Crab_2004_rmf.fits.gz for Source 1
Using Auxiliary Response (ARF) File Crab_2004_arf.fits.gz
Fit statistic : Chi-Squared -nan using 62 bins.
Test statistic : Chi-Squared -nan using 62 bins.
Current data and model not fit yet.
ignore: 1 channels ignored from source number 1
Fit statistic : Chi-Squared -nan using 61 bins.
Test statistic : Chi-Squared -nan using 61 bins.
Current data and model not fit yet.
2 channels (61-62) ignored in spectrum # 1
Fit statistic : Chi-Squared -nan using 59 bins.
Test statistic : Chi-Squared -nan using 59 bins.
Current data and model not fit yet.
8 channels (1-8) ignored in spectrum # 1
2 channels (61-62) ignored in spectrum # 1
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared 16610.24 using 52 bins.
Test statistic : Chi-Squared 16610.24 using 52 bins.
Null hypothesis probability of 0.00e+00 with 47 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 16610.24 using 52 bins.
Test statistic : Chi-Sqlg10Fluxuared 16610.24 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 16610.24 using 52 bins.
Test statistic : Chi-Squared 16610.24 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 17625.68 using 52 bins.
Test statistic : Chi-Squared 17625.68 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 16824.81 using 52 bins.
Test statistic : Chi-Squared 16824.81 using 52 bins.
Null hypothesis probability of 0.00e+00 with 49 degrees of freedom
Current data and model not fit yet.
Warning: renorm - no variable model to allow renormalization
Parameters
Chi-Squared |beta|/N Lvl 3:lg10Flux 4:PhoIndx1 6:PhoIndx2
1223.69 27995.4 -3 -7.79892 2.09392 2.34579
43.6611 11589.6 -4 -7.83234 2.06677 2.31150
41.3724 459.142 -5 -7.83389 2.06456 2.31103
41.3723 3.36647 -6 -7.83390 2.06455 2.31103
========================================
Variances and Principal Axes
3 4 6
1.1051E-06| 0.9998 -0.0191 -0.0077
2.3880E-05| 0.0195 0.9980 0.0597
1.4345E-03| -0.0065 0.0599 -0.9982
----------------------------------------
====================================
Covariance Matrix
1 2 3
1.175e-06 -1.148e-07 9.347e-06
-1.148e-07 2.893e-05 -8.430e-05
9.347e-06 -8.430e-05 1.429e-03
------------------------------------
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 20.0000 frozen
2 1 cflux Emax keV 80.0000 frozen
3 1 cflux lg10Flux cgs -7.83390 +/- 1.08378E-03
4 2 bknpower PhoIndx1 2.06455 +/- 5.37847E-03
5 2 bknpower BreakE keV 100.000 frozen
6 2 bknpower PhoIndx2 2.31103 +/- 3.78077E-02
7 2 bknpower norm 1.00000 frozen
________________________________________________________________________
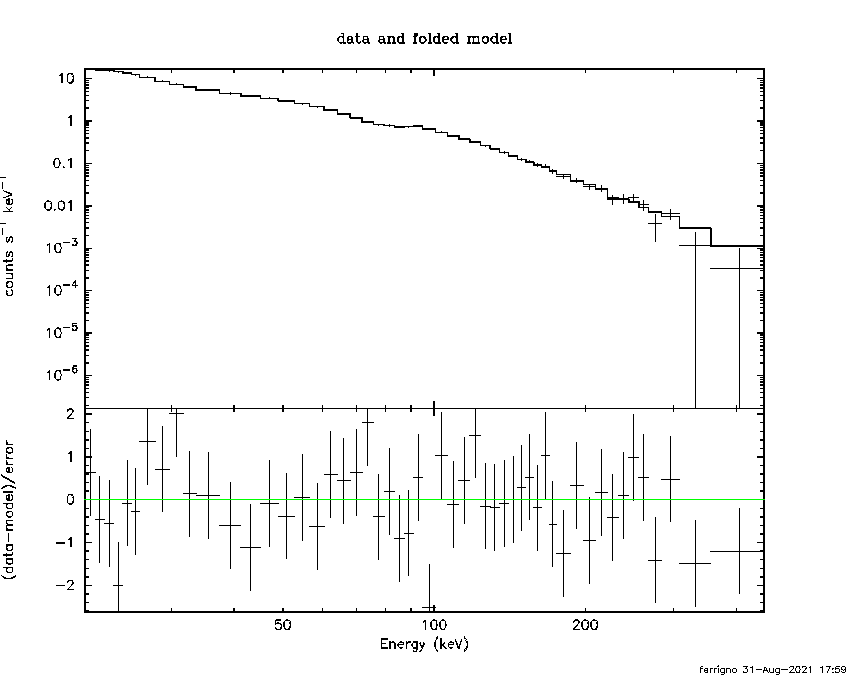
Fit statistic : Chi-Squared 41.37 using 52 bins.
Test statistic : Chi-Squared 41.37 using 52 bins.
Null hypothesis probability of 7.72e-01 with 49 degrees of freedom
parameter 5 is already frozen.
Warning: renorm - no variable model to allow renormalization
Parameters
Chi-Squared |beta|/N Lvl 3:lg10Flux 4:PhoIndx1 6:PhoIndx2
41.3723 0.0183563 -3 -7.83390 2.06455 2.31103
========================================
Variances and Principal Axes
3 4 6
1.1051E-06| 0.9998 -0.0191 -0.0077
2.3882E-05| 0.0195 0.9980 0.0597
1.4346E-03| -0.0065 0.0599 -0.9982
----------------------------------------
====================================
Covariance Matrix
1 2 3
1.175e-06 -1.148e-07 9.347e-06
-1.148e-07 2.893e-05 -8.431e-05
9.347e-06 -8.431e-05 1.429e-03
------------------------------------
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 20.0000 frozen
2 1 cflux Emax keV 80.0000 frozen
3 1 cflux lg10Flux cgs -7. 83390 +/- 1.08381E-03
4 2 bknpower PhoIndx1 2.06455 +/- 5.37861E-03
5 2 bknpower BreakE keV 100.000 frozen
6 2 bknpower PhoIndx2 2.31103 +/- 3.78086E-02
7 2 bknpower norm 1.00000 frozen
________________________________________________________________________
Fit statistic : Chi-Squared 41.37 using 52 bins.
Test statistic : Chi-Squared 41.37 using 52 bins.
Null hypothesis probability of 7.72e-01 with 49 degrees of freedom
*** Parameter 1 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 2 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 5 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 7 is not a variable model parameter and no confidence range will be calculated.
Parameter Confidence Range (-7.83 False False
PhoIndx1 2.06 False False
PhoIndx2 2.31 False False
1)
3 -7.83499 -7.83281 (-0.0010919,0.00108657)
4 2.05918 2.06992 (-0.00536723,0.00537103)
6 2.27383 2.34882 (-0.0372053,0.0377893)
***Warning: POISSERR keyword is missing or of wrong format, assuming FALSE.
PGPLOT /png: writing new file as pgplot.png_2
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/Off
Model Model Component Parameter Unit Value
par comp
7
1 1 cflux Emin keV 0.500000 frozen
2 1 cflux Emax keV 10.0000 frozen
3 1 cflux lg10Flux cgs -12.0000 +/- 0.0
4 2 bknpower PhoIndx1 1.00000 +/- 0.0
5 2 bknpower BreakE keV 5.00000 +/- 0.0
6 2 bknpower PhoIndx2 2.00000 +/- 0.0
7 2 bknpower norm 1.00000 +/- 0.0
________________________________________________________________________
Warning: RMF CHANTYPE keyword (PHA) is not consistent with that from spectrum (PI)
1 spectrum in use
Spectral Data File: Crab_2005_spectrum.fits Spectrum 1
Net count rate (cts/s) for Spectrum:1 -nan +/- -nan
Assigned to Data Group 1 and Plot Group 1
Noticed Channels: 1-62
Telescope: INTEGRAL Instrument: IBIS Channel Type: PI
Exposure Time: 1.879e+04 sec
Using fit statistic: chi
Using Response (RISGRI ignore: **-20.00,500.-**MF) File Crab_2005_rmf.fits.gz for Source 1
Using Auxiliary Response (ARF) File Crab_2005_arf.fits.gz
Fit statistic : Chi-Squared -nan using 62 bins.
Test statistic : Chi-Squared -nan using 62 bins.
Current data and model not fit yet.
ignore: 1 channels ignored from source number 1
Fit statistic : Chi-Squared -nan using 61 bins.
Test statistic : Chi-Squared -nan using 61 bins.
Current data and model not fit yet.
2 channels (61-62) ignored in spectrum # 1
Fit statistic : Chi-Squared -nan using 59 bins.
Test statistic : Chi-Squared -nan using 59 bins.
Current data and model not fit yet.
***Warning: POISSERR keyword is missing or of wrong format, assuming FALSE.
8 channels (1-8) ignored in spectrum # 1
2 channels (61-62) ignored in spectrum # 1
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared 19542.13 using 52 bins.
Test statistic : Chi-Squared 19542.13 using 52 bins.
Null hypothesis probability of 0.00e+00 with 47 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 19542.13 using 52 bins.
Test statistic : Chi-Sqlg10Flux -7.82 False False
PhoIndx1 2.19 False False
PhoIndx2 2.47 False False
uared 19542.13 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 19542.13 using 52 bins.
Test statistic : Chi-Squared 19542.13 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 20464.06 using 52 bins.
Test statistic : Chi-Squared 20464.06 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 19707.72 using 52 bins.
Test statistic : Chi-Squared 19707.72 using 52 bins.
Null hypothesis probability of 0.00e+00 with 49 degrees of freedom
Current data and model not fit yet.
Warning: renorm - no variable model to allow renormalization
Parameters
Chi-Squared |beta|/N Lvl 3:lg10Flux 4:PhoIndx1 6:PhoIndx2
2262.53 29389.4 -3 -7.77822 2.28458 2.48308
237.783 14465.1 -4 -7.81808 2.19880 2.46927
231.336 673.729 -5 -7.82039 2.19156 2.47344
231.336 5.96194 -6 -7.82041 2.19156 2.47344
========================================
Variances and Principal Axes
3 4 6
1.0358E-06| 0.9990 -0.0435 -0.0093
2.1151E-05| -0.0440 -0.9969 -0.0657
1.1917E-03| 0.0064 -0.0660 0.9978
----------------------------------------
====================================
Covariance Matrix
1 2 3
1.124e-06 3.785e-07 7.668e-06
3.785e-07 2.621e-05 -7.709e-05
7.668e-06 -7.709e-05 1.187e-03
------------------------------------
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 20.0000 frozen
2 1 cflux Emax keV 80.0000 frozen
3 1 cflux lg10Flux cgs -7.82041 +/- 1.05997E-03
4 2 bknpower PhoIndx1 2.19156 +/- 5.11975E-03
5 2 bknpower BreakE keV 100.000 frozen
6 2 bknpower PhoIndx2 2.47344 +/- 3.44470E-02
7 2 bknpower norm 1.00000 frozen
________________________________________________________________________
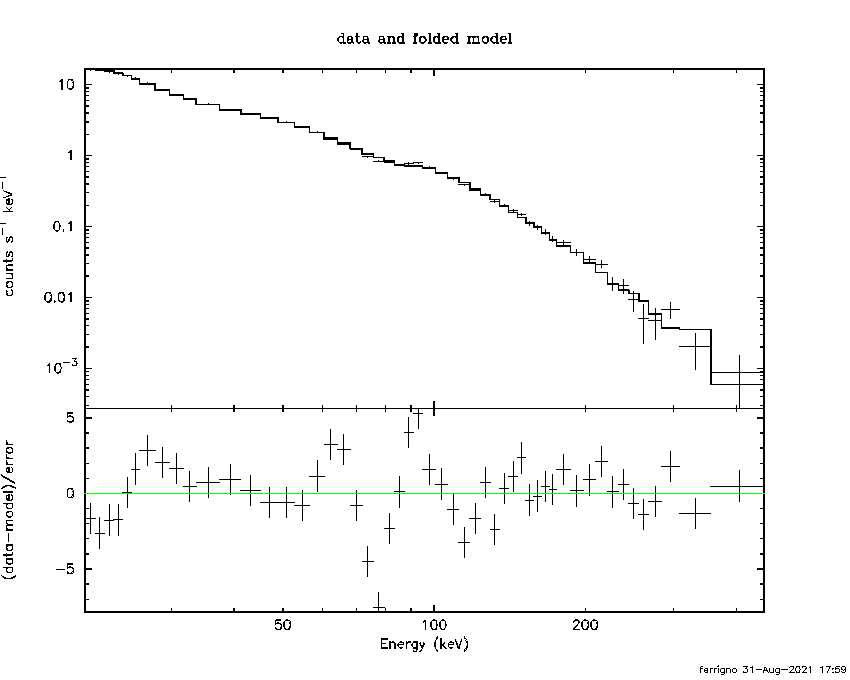
Fit statistic : Chi-Squared 231.34 using 52 bins.
Test statistic : Chi-Squared 231.34 using 52 bins.
Null hypothesis probability of 1.77e-25 with 49 degrees of freedom
parameter 5 is already frozen.
Warning: renorm - no variable model to allow renormalization
Parameters
Chi-Squared |beta
PGPLOT /png: writing new file as pgplot.png_2
***Warning: POISSERR keyword is missing or of wrong format, assuming FALSE.
|/N Lvl 3:lg10Flux 4:PhoIndx1 6:PhoIndx2
231.336 0.032211 -3 -7.82041 2.19156 2.47344
========================================
Variances and Principal Axes
3 4 6
1.0359E-06| 0.9990 -0.0435 -0.0093
2.1153E-05| -0.0440 -0.9969 -0.0657
1.1918E-03| 0.0064 -0.0660 0.9978
----------------------------------------
====================================
Covariance Matrix
1 2 3
1.124e-06 3.785e-07 7.669e-06
3.785e-07 2.621e-05 -7.710e-05
7.669e-06 -7.710e-05 1.187e-03
------------------------------------
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 20.0000 frozen
2 1 cflux Emax keV 80.0000 frozen
3 1 cflux lg10Flux cgs -7.82041 +/- 1.06002E-03
4 2 bknpower PhoIndx1 2.19156 +/- 5.11997E-03
5 2 bknpower BreakE keV 100.000 frozen
6 2 bknpower PhoIndx2 2.47344 +/- 3.44484E-02
7 2 bknpower norm 1.00000 frozen
________________________________________________________________________
Fit statistic : Chi-Squared 231.34 using 52 bins.
Test statistic : Chi-Squared 231.34 using 52 bins.
Null hypothesis probability of 1.77e-25 with 49 degrees of freedom
*** Parameter 1 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 2 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 5 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 7 is not a variable model parameter and no confidence range will be calculated.
Parameter Confidence Range (1)
3 -7.82148 -7.81935 (-0.00106875,0.00106364)
4 2.18645 2.19668 (-0.00511457,0.00511685)
6 2.43891 2.50843 (-0.0345338,0.034985)
7
ISGRI ignore: **-20.00,500.-**
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/Off
lg10Flux Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 0.500000 frozen
2 1 cflux Emax keV 10.0000 frozen
3 1 cflux lg10Flux cgs -12.0000 +/- 0.0
4 2 bknpower PhoIndx1 1.00000 +/- 0.0
5 2 bknpower BreakE keV 5.00000 +/- 0.0
6 2 bknpower PhoIndx2 2.00000 +/- 0.0
7 2 bknpower norm 1.00000 +/- 0.0
________________________________________________________________________
Warning: RMF CHANTYPE keyword (PHA) is not consistent with that from spectrum (PI)
1 spectrum in use
Spectral Data File: Crab_2006_spectrum.fits Spectrum 1
Net count rate (cts/s) for Spectrum:1 -nan +/- -nan
Assigned to Data Group 1 and Plot Group 1
Noticed Channels: 1-62
Telescope: INTEGRAL Instrument: IBIS Channel Type: PI
Exposure Time-7.83 False False
PhoIndx1 2.08 False False
PhoIndx2 2.26 False : 1.455e+04 sec
Using fit statistic: chi
Using Response (RMF) File Crab_2006_rmf.fits.gz for Source 1
Using Auxiliary Response (ARF) File Crab_2006_arf.fits.gz
Fit statistic : Chi-Squared -nan using 62 bins.
Test statistic : Chi-Squared -nan using 62 bins.
Current data and model not fit yet.
ignore: 1 channels ignored from source number 1
Fit statistic : Chi-Squared -nan using 61 bins.
Test statistic : Chi-Squared -nan using 61 bins.
Current data and model not fit yet.
2 channels (61-62) ignored in spectrum # 1
Fit statistic : Chi-Squared -nan using 59 bins.
Test statistic : Chi-Squared -nan using 59 bins.
Current data and model not fit yet.
8 channels (1-8) ignored in spectrum # 1
2 channels (61-62) ignored in spectrum # 1
Fit statistic : Chi-Squared -nan using 52False
bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared -nan using 52 bins.
Test statistic : Chi-Squared -nan using 52 bins.
Current data and model not fit yet.
Fit statistic : Chi-Squared 17003.74 using 52 bins.
Test statistic : Chi-Squared 17003.74 using 52 bins.
Null hypothesis probability of 0.00e+00 with 47 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 17003.74 using 52 bins.
Test statistic : Chi-Squared 17003.74 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 17003.74 using 52 bins.
Test statistic : Chi-Squared 17003.74 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 18020.00 using 52 bins.
Test statistic : Chi-Squared 18020.00 using 52 bins.
Null hypothesis probability of 0.00e+00 with 48 degrees of freedom
Current data and model not fit yet.
Fit statistic : Chi-Squared 17244.66 using 52 bins.
Test statistic : Chi-Squared 17244.66 using 52 bins.
Null hypothesis probability of 0.00e+00 with 49 degrees of freedom
Current data and model not fit yet.
Warning: renorm - no variable model to allow renormalization
Parameters
Chi-Squared |beta|/N Lvl 3:lg10Flux 4:PhoIndx1 6:PhoIndx2
1260.99 28621.1 -3 -7.79884 2.11203 2.26811
32.4047
PGPLOT /png: writing new file as pgplot.png_2
11903.5 -4 -7.83229 2.07958 2.25587
29.9577 473.3 -5 -7.83386 2.07700 2.25598
29.9576 3.49072 -6 -7.83387 2.07699 2.25598
========================================
Variances and Principal Axes
3 4 6
1.0804E-06| 0.9998 -0.0198 -0.0083
2.2834E-05| 0.0202 0.9978 0.0633
1.3097E-03| -0.0070 0.0635 -0.9980
----------------------------------------
====================================
Covariance Matrix
1 2 3
1.154e-06 -1.446e-07 9.210e-06
-1.446e-07 2.801e-05 -8.153e-05
9.210e-06 -8.153e-05 1.304e-03
------------------------------------
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 20.0000 frozen
2 1 cflux Emax keV 80.0000 frozen
3 1 cflux lg10Flux cgs -7.83387 +/- 1.07426E-03
4 2 bknpower PhoIndx1 2.07699 +/- 5.29259E-03
5 2 bknpower BreakE keV 100.000 frozen
6 2 bknpower PhoIndx2 2.25598 +/- 3.61174E-02
7 2 bknpower norm 1.00000 frozen
________________________________________________________________________
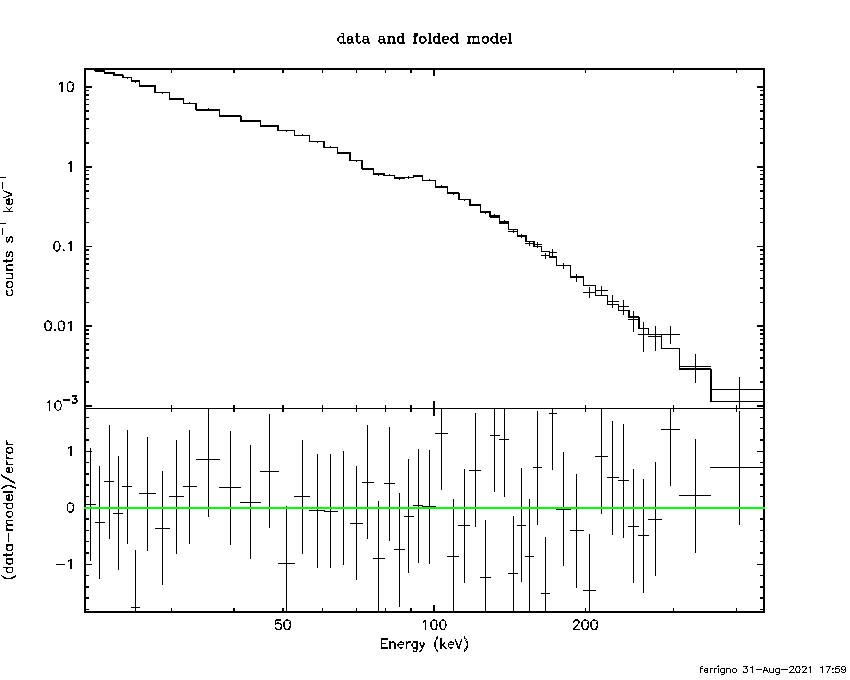
Fit statistic : Chi-Squared 29.96 using 52 bins.
Test statistic : Chi-Squared 29.96 using 52 bins.
Null hypothesis probability of 9.85e-01 with 49 degrees of freedom
parameter 5 is already frozen.
Warning: renorm - no variable model to allow renormalization
Parameters
Chi-Squared |beta|/N Lvl 3:lg10Flux 4:PhoIndx1 6:PhoIndx2
29.9576 0.0191331 -3 -7.83387 2.07699 2.25598
========================================
Variances and Principal Axes
3 4 6
1.0805E-06| 0.9998 -0.0198 -0.0083
2.2835E-05| 0.0202 0.9978 0.0633
1.3098E-03| -0.0070 0.0635 -0.9980
----------------------------------------
====================================
Covariance Matrix
1 2 3
1.154e-06 -1.446e-07 9.210e-06
-1.446e-07 2.801e-05 -8.153e-05
9.210e-06 -8.153e-05 1.305e-03
------------------------------------
========================================================================
Model cflux<1>*bknpower<2> Source No.: 1 Active/On
Model Model Component Parameter Unit Value
par comp
1 1 cflux Emin keV 20.0000 frozen
2 1 cflux Emax keV 80.0000 frozen
3 1 cflux lg10Flux cgs -7.83387 +/- 1.07429E-03
4 2 bknpower PhoIndx1 2.07699 +/- 5.29272E-03
5 2 bknpower BreakE keV 100.000 frozen
6 2 bknpower PhoIndx2 2.25598 +/- 3.61181E-02
7 2 bknpower norm 1.00000 frozen
________________________________________________________________________
Fit statistic : Chi-Squared 29.96 using 52 bins.
Test statistic : Chi-Squared 29.96 using 52 bins.
Null hypothesis probability of 9.85e-01 with 49 degrees of freedom
*** Parameter 1 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 2 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 5 is not a variable model parameter and no confidence range will be calculated.
*** Parameter 7 is not a variable model parameter and no confidence range will be calculated.
Parameter Confidence Range (1)
3 -7.83496 -7.8328 (-0.00108288,0.00107766)
4 2.0717 2.08229 (-0.00529128,0.00529666)
6 2.22004 2.29245 (-0.0359406,0.0364654)